DNA Whole Genome Sequencing - astrology for sigmas
I discovered that I couldn't find comprehensive guides on genetic testing, so I wrote one.
Update:
September 9, 2024:
Updated information on high-effect variants and polygenic risk score. Highly recommended for reading.
Updated information on Sequencing.com and Nebula
Added information about Genetic Lifehacks and Self-decode
Introduction
In July, I took a Whole Genome Sequencing DNA test.
After receiving the results, I realized that no one had shared their experience of taking the test.
Also, nobody had shared which third-party services you can use to get more information about your health, ancestry, and traits.
Therefore, I decided to write this blog post to make it easier for you to decide whether to take a DNA test and to help you extract more useful information from it.
DNA 101
Proteins are the main workers of our organism, responsible for all processes—muscle contraction, sugar digestion, thinking process.
DNA contains instructions for creating the proteins in our body.
Identical DNA is stored in the nucleus of EVERY cell in chromosomes.
Humans have 23 pairs of chromosomes.
23 chromosomes are received from the father and 23 chromosomes from the mother.
Autosomes
The first 22 chromosomes are called autosomal.
Autosomal DNA is passed down to us from both mom and dad. It's transmitted in roughly equal proportions.
Consequently, each gene usually has two versions (alleles) - one from mom, the other from dad, which allows us to hedge risks - if the protein from one gene doesn't work, the protein from the second will still function.
Homozygosity vs Heterozygosity
A mutation is homozygous if it's present in both alleles inherited from parents.
A mutation is called heterozygous if it's present in only one allele inherited from parents.
Autosomal Recessive Traits vs Autosomal Dominant Traits
Autosomal recessive traits manifest if a certain mutation is activated only when it's homozygous - i.e., all versions of the gene must have the mutation.
Autosomal dominant traits manifest if a certain mutation is activated when it's heterozygous or homozygous - i.e., there must be at least one gene with the mutation for it to manifest.
Sex Chromosomes (23rd Pair)
The 23rd chromosome differs between men and women.
Men have an XY pair, while women have an XX pair.
This gives rise to some interesting properties:
A woman always passes an X chromosome, so she doesn't determine the child's sex.
A man's sperm can have either the father's X chromosome, resulting in an XX set and a girl, or a Y chromosome, resulting in an XY set and a boy.
We can only get the Y chromosome from our father, who could only get it from his father, and so on.
Thus, our Y chromosome will be very similar to the Y chromosome of our father, his father's father, and so on.
Mitochondrial DNA
Each of our cells has mitochondria, ancient bacteria that once entered our cells and turned out to be awesome symbionts. Mitochondria help us create universal ATP batteries from all the horrors we've eaten.
Mitochondria also have DNA inside them. Since Mitochondrial DNA is only passed from the egg cell, Mitochondrial DNA is only inherited from the mother.
Our mtDNA will be similar to the DNA of our mother, and our mother's mother, and so on.
Whole Genome Sequencing vs Genotyping
23andMe, Ancestry, MyHeritage, and other popular tests are genotyping tests. They scan only 0.1% of all the DNA you have.
WGS (Whole Genome Sequencing) differs in that, unlike 23andMe, Ancestry, and others, you get your entire genome - ≈100%.
Advantages of Whole Genome Sequencing:
We get more information from the genome.
If a new study emerges revealing dangerous mutations in "unpopular" genes, re-sequencing won't be necessary, as you already have the entire genome.
The full genome can be converted into all genotyping formats, as genotyping formats are just parts of the full genome recorded in a special way. This allows you to upload your genome to all services requiring data from 23andMe, Ancestry, MyHeritage, and other providers.
On advice, I chose Nucleus. WGS from Nucleus costs only $400. What a catch!
WGS alternatives to Nucleus
I discovered that not only Nucleus does Whole Genome Sequencing now, but also about a dozen other companies.
Some at prices similar to Nucleus's.
Nucleus is currently only available in America, so we ordered a test from Nebula for my brother in Latvia.
Here's a list of alternatives, I recommend checking it out:
Taking the test
A week after purchase, my test arrived!
Beautiful packaging:
Beautiful inside:
Taking it is not difficult.
You follow the instructions:
collect saliva in a special container
pack the container in a special bag
put the bag in a postal envelope
drop the postal envelope in any USPS mailbox.
Easy-peasy.
On August 5th, the guys received my DNA by mail. By the 26th, the results were ready in 3 weeks. That's twice as fast as expected.
W for Nucleus.
> My friend decided to take the same test 5 days later, but his answer came at the same time as mine. Apparently, they do sequencing in batches.
Results
Information from the DNA test can be divided into three types:
Health - what diseases you have a genetic predisposition to
Ancestry - where your ancestors come from
Traits / IQ - your genetic predisposition to certain behavioral or physical traits
Below, I compared the results of various services with the baseline and with each other.
Health
Polygenic risk score, High-effect variants and Genetic Variant Functional Impact
In this section, I will describe how we can assess the impact of variations in the genome on human health.
To predict predisposition to diseases, two things are considered: High-effect variants and Polygenic risk score.
Probability of variation pathogenicity
Usually, to calculate how much a gene change has affected protein function, metrics like CADD are used.
The higher the CADD score, the more the protein function has changed.
As a rule of thumb, only consider variations with a CADD score greater than 20.
It's important to understand that CADD only calculates the probability that the protein will function differently.
CADD does not indicate whether there is a link between the altered protein and the disease.
Also, CADD does not indicate how significantly a specific protein affects the disease.
High-effect variants
High-effect variants - when ONE mutation in DNA significantly increases predisposition to a disease.
Variants associated with a disease can be identified by serious providers like Nucleus and Nebula.
Additionally, you can try to find such mutations on your own using Variant Exploring services like GenVue:
Look for mutations with a high CADD score (>20).
Find out which diseases are associated with this mutation.
Study an article or ask ChatGPT about the variation.
Learn how and to what extent this variation affects gene function and how much it increases the chance of potential disease.
Remember, CADD does not indicate how much a mutation increases your predisposition to a specific disease.
CADD only indicates how the protein's function will be altered due to the variation.
Polygenic risk score (PRS)
Polygenic risk score - when a COMBINATION of DNA mutations increases predisposition to a disease.
Usually, small mutations do not significantly increase predisposition to a disease, but their combination can sometimes significantly increase the impact.
PRS calculates how the combination of mutations increases the chance of developing a disease.
Services like Nucleus and Nebula can calculate PRS.
Calculating PRS on your own
Calculating PRS on your own is difficult and requires data scientist skills.
Here's an example of how Nucleus calculates it:
I haven't tried it myself yet, but it seems like a solvable task.
If you attempt it, please let me know!
Single common-effect variant mutations
Many providers offer conclusions based on one or a few mutations that are not high-effect mutations.
Fatal mistake!
These mutations alone have too little power to have a negative impact, so it's not worth looking at or worrying about.
Here's a comment from the CEO of Nucleus on this topic:
Searching for High-effect variants on your own
Nucleus
Price: $49/year
Nucleus itself analyzes DNA and allows the user to look at already calculated predispositions to diseases. It tests only 22 diseases.
It will point out both high-effect variants and polygenic risk score.
Here's an example of what data you'll get for each ailment:
For polygenic risk score, it calculates:
the overall chance that I will get the disease during my lifetime
Z-score - and in which direction my result differs from the standard distribution
comparison of my risk with the risk of various groups of people
At the end, data for each disease can be downloaded as a report that can be shown to a doctor.
List of what Nucleus currently tests for
ADHD
Age-related macular degeneration
Alcohol dependence
Alzheimer's disease
Anxiety disorders
Asthma
Bipolar disorder
Celiac disease
Colorectal cancer
Coronary artery disease
Depression
Gastric cancer
Hypertension
Insomnia
Male breast cancer
Migraine
Multiple sclerosis
OCD
Parkinson's disease
Prostate cancer
Rheumatoid arthritis
Schizophrenia
Type 2 diabetes
Conclusion
Among the advantages - a huge list of polygenic risk score information, it's extremely difficult to find and very few sources provide it.
Everything is written in understandable language, the absolute chance of a certain disease is clear and how much higher it is than the population average.
With Nucleus, I found a crazy predisposition to one disease (only 0.0082% of people have a higher predisposition to the disease than me). This ailment was observed in my family.
Now the plan is to control disease markers, as well as change daily diets.
One downside was the lack of interpretability of the provided PRS results; I wanted to know:
Which variations in which genes have the most impact on a specific ailment?
What biomarkers should I be looking at?
What should be done to mitigate the risk of developing the disease?
I had to use other services for assistance.
GeneticGenie
Price: Free
GenVue accepts WGS data in VCF format—you can download them from the Nucleus website or your provider.
GeneticGenie has three functions:
Methylation panel
Detox panel
GenVue
Methylation and Detox panels
First, I recommend looking at the results of Genomic Panels already collected for us.
Currently, there are two panels:
Methylation panel - methylation controls how many proteins will be created in what quantity
Detox panel - detoxification removes toxins from the body
My pipeline was:
Upload VCF file
Get results from the panel
Copy the results to ChatGPT and ask for an explanation.
ChatGPT interprets the results fairly well, but for quality control, simultaneously check if the model is hallucinating.
If you want to draw any important conclusions from what you have found, be sure to read articles and consult with a doctor and Reddit.
So far, ChatGPT is extremely erudite in gene mutations and so far I've been hallucinating more than it has.
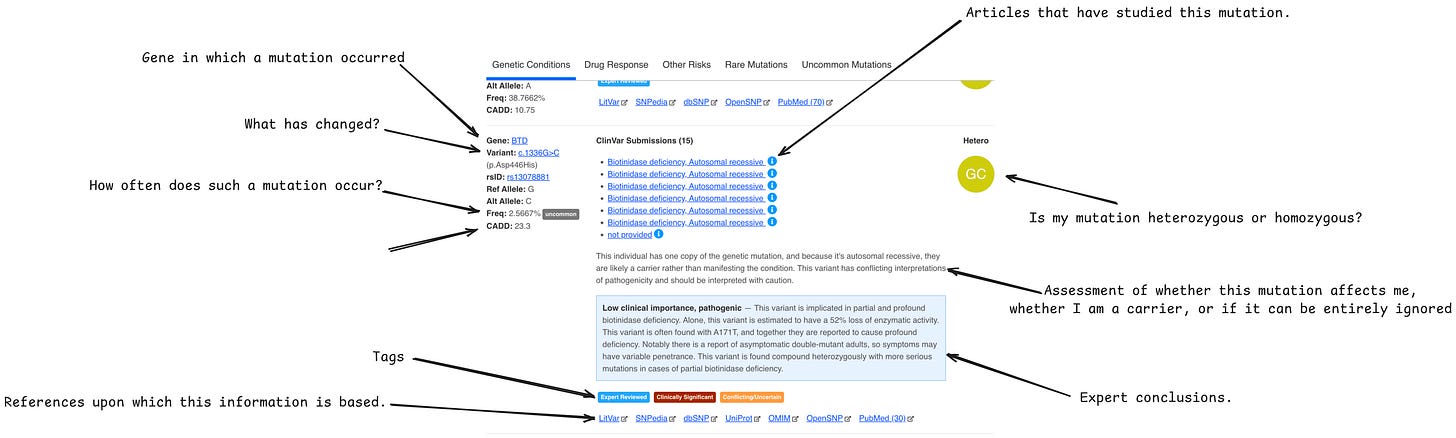
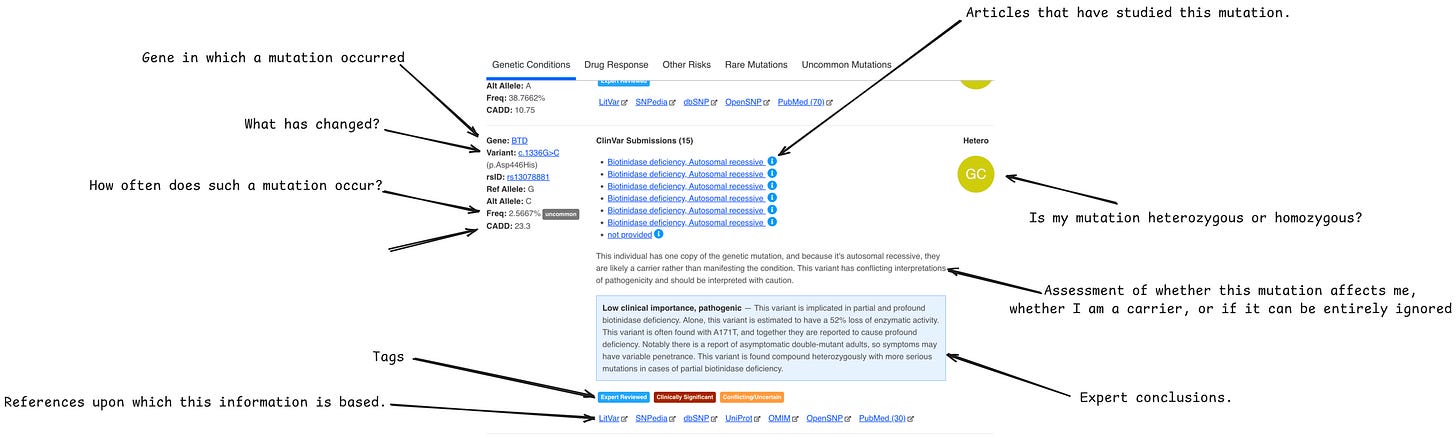
GenVue
This is already a more serious scanner of single DNA variations.
It gives 5 types of results:
genetic conditions - mutations have been reviewed by an expert and affect a person's well-being
drug response - mutations have been reviewed by an expert and affect susceptibility to drugs
other risks - mutations that have been noted as clinically important but not verified by an expert
rare mutations - mutations that occur with a chance of less than 1%
uncommon mutations - mutations that occur with a chance from 1% to 5%
The image above shows the GenVue interface.
I found that the first two tabs are the most important:
In the first one, we look at high-effective genes
In the second one, we save wherever our reactions to which drugs will be.
Be sure to check CADD >20. Also, in the expert comments, there is often very valuable information that I haven't seen anywhere else except in GenVue.
Conclusion
This is the second health editor that has proven to be truly useful.
Deviation is clear, risk is understood, there are links to articles, it's clear what to do.
Based on this data, it is clear which expert to consult and which biomarkers to look at.
It complements Nucleus' results excellently.
Nebula
Link
Pricing: Free Trial (But you must manage to cancel after the report is generated, but before the free trial ends.)
In addition to offering its WGS tests, Nebula allows you to upload data in the format of 23andMe or Ancestry to obtain results (for instructions on how to convert your genome into Genotyping formats, see the Appendix).
Nebula returns Polygenic Risk Score associated with specific diseases in the results. Additionally, Nebula shows traits, which will be described in the traits chapter below.
Sketchy subscription thing
While your DNA Report is still being compiled, you cannot cancel the subscription.
Once it is ready, you will be able to cancel it.
Results
Nebula, like Nucleus, informs about diseases and in what percentile of predisposition to the disease I stand.
Based on Genotyping data, Nebula was able to provide PRS for 10 rare diseases.
I believe that if WGS is submitted to Nebula, the number of diseases for which PRS will be calculated would be greater than that of Nucleus.
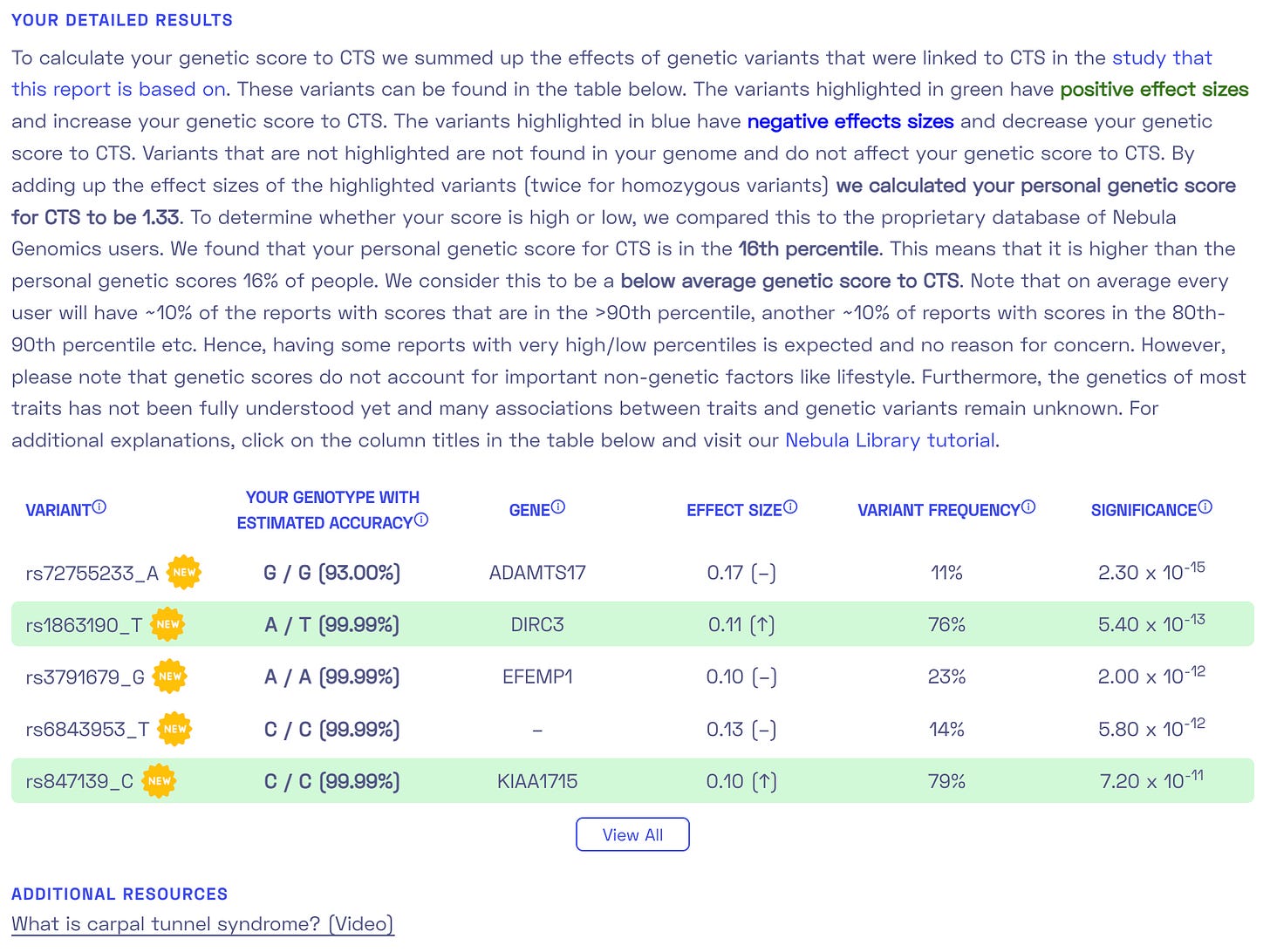
I liked that Nebula has a clear open report on which variations and how much they influenced the final PRS.
Unfortunately, unlike Nucleus, there is no information on how much my chances of having a specific ailment increase or decrease compared to the average person.
Also, there are no subsequent instructions on which biomarkers to watch for, and what activities should and should not be done.
Conclusion
Nebula is the second service using polygenic risk score.
Unlike Nucleus, the reports are less detailed, but I assume that when WGS is submitted here, there would be more.
Since Nucleus does not operate in Europe, I ordered Nebula for my girlfriend and brother, I will write about their experience with it.
Genetic Lifehacks
Link
Price: $10 monthly/ $50 annually (Articles are free!)
A reader of this post recommended Genetic Lifehacks to me, for which I am very grateful!
Genetic Lifehacks accepts Genotyping data.
Based on this data, reports are generated for a vast number of categories:
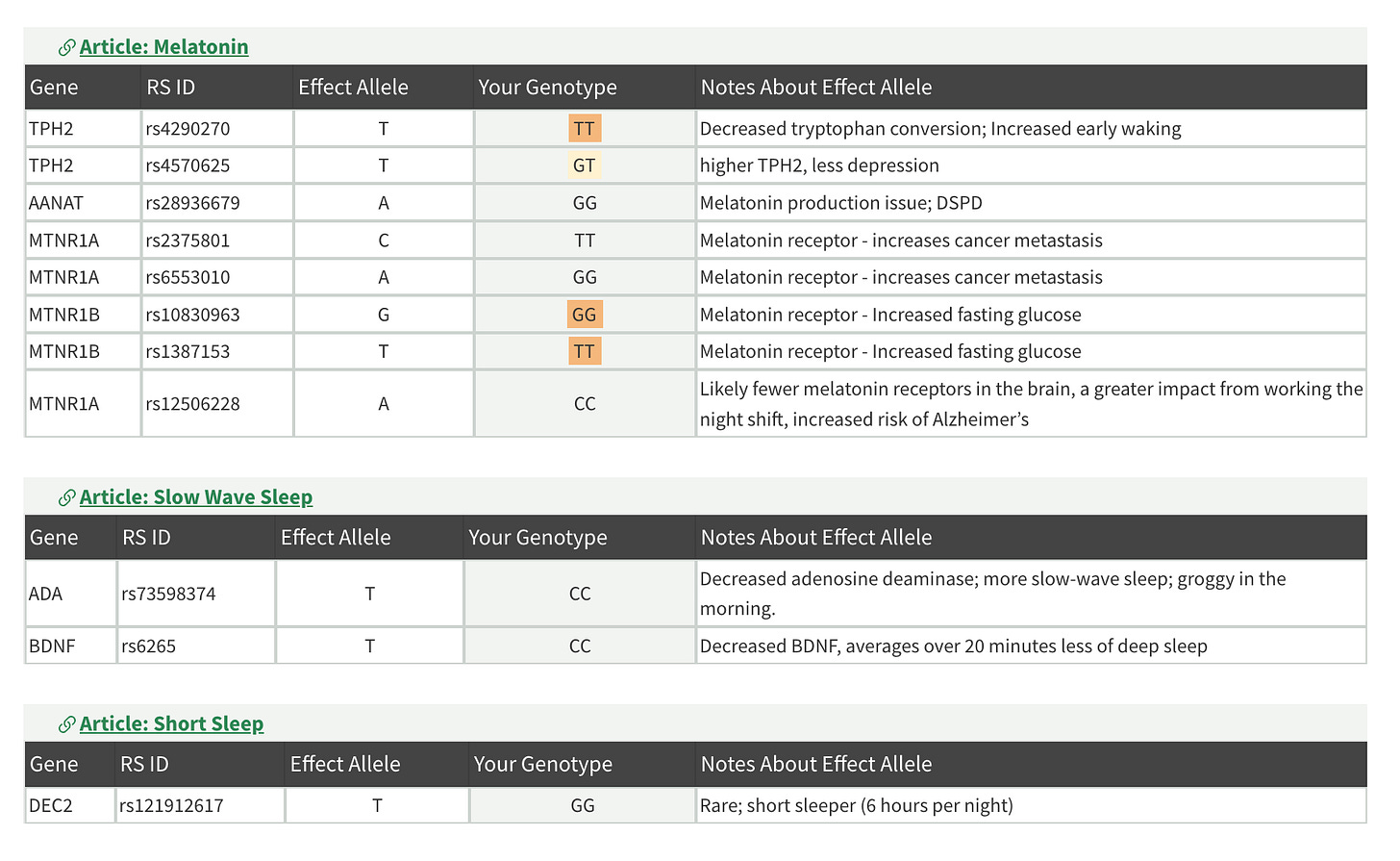
For example, reports of this kind in the Sleep&Circadian category:
For each report on the Genetic Lifehacks website, there is an article that intricately describes the workings of the biological system:
If you have Premium, the article will also provide a detailed interpretation of your variations:
Conclusion
Looks very cool:
Reports are grouped by topics.
Each report is accompanied by an extremely detailed article.
Each article includes links to papers, as well as a detailed explanation of which variation I have.
It's a shame that this service currently does not accept WGS files.
Also, for most reports, it is not stated how much a variation increases my predisposition in quantitative terms.
Does the risk of diabetes increase by 1% or by two times?
In my humble opinion, this is very important for decision-making.
Sequencing.com
Pricing: $39/mo (there are alternative plans, but you don't need them)
Sequencing.com is a huge platform for WGS analysis.
They provide:
Next-Gen Disease Screen - a detailed explorer of variations that affect health
HealthScan - a less detailed explorer of variations that affect health (I didn't understand how it differs from Next-Gen Disease Screen)
SequencingAI - an LLM to interact with to understand the interpretation of results
Shop - in the store, you can buy third-party plugins for DNA analysis
Data upload
Since we want to upload the entire genome, sequencing.com requires doing this through their special program "Big Yotta".
On my MacOS, this program simply wouldn't launch.
I reported this to sequencing, hopefully they'll fix it soon.
I managed to launch Big Yotta on Windows and upload the VCF file through it.
Next-Gen Disease Screen
The interface is similar to GenVue.
But here conditions are divided by risk size, which is easier to navigate.
They tell you what conditions you have, how dangerous they are, and how confident sequencing is that this condition arises due to a specific change.
I missed the expert comments that allowed for a deeper scientific conclusion like in GenVue.
For example, one mutation was found by both GenVue and Next-Gen Disease Screen.
In GenVue, there was also a comment that despite this disease only being present in those who have the current gene variation, the condition has low penetrance - accordingly, it's unlikely to develop it. There was nothing like this in Sequencing.com.
It was a bit harder to separate the wheat from the chaff than in GenVue.
HealthScan
I didn't understand what the difference is between Next-Gen Disease and HealthScan. It's as if HealthScan just has fewer variations.
Sometimes the condition names are incorrect:
Shop
The store provides endless tests from other providers.
The tests are anomalously expensive, I decided it's not worth it.
There are also free tests:
Am I a joke to you???
Conclusions
Sequencing.com is clearly not worth paying $468 annually for.
It's better to just use free GenVue.
Waiting for HealthScan results. Maybe they'll make me change my mind.
Other Health tests
Nebula
Link
Pricing: Free Trial (But you can cancel it, only after DNA report is generated)
Nebula is a bit strange. It offers uploading your DNA to get results through a Free Trial.
But here's what the subscription settings look like after the free trial was set up.
There's no DNA report either:
Support explained to me that the free trial will start when nebula generates the DNA report and only then can I cancel this free trial. Sounds sketchy...
Considering how many people use and recommend Nebula, this was a surprise to me.
I hope the guys fix the interface bugs.
Since Nebula is the most popular WGS, I ordered it for my girlfriend and brother, I'll write about what experience they have with getting the genome.
BioCodify
Link
Pricing: $15/year
Biocodify allows you to look at variations in the genome and conditions that come from it.
Functionality is worse than GenVue + costs money.
It was difficult for me to separate the wheat from the chaff. To understand what's really worth studying and what's an unstudied rare risk.
For now, I don't recommend using it, but I would closely follow this project, as the developers are actively developing it and listening to user experience.
Accordingly, there's every chance to influence and make a cool product.
Sophisticated tools
There are https://gene.iobio.io/ and https://run.opencravat.org/
which also provide some variance analysis, but these tools are too sophisticated. I decided not to go down the rabbit hole, but you can.
Please report your results!
An acquaintance geneticist informed me that they use https://github.com/broadinstitute/seqr for analyzing rare diseases.
Self-decode
Link
Price: $200 first year, then $100 annually
I've been advised a couple of times to try self-decode for polygenic risk score.
It costs $200 per year, so I've decided not to try it for now.
If you have used it, please share your results!
General conclusion on health
Nucleus, GeneVue, Genetic Health, and Nebula provided the real signal without noise.
I would like to find a provider with high-quality polygenic risk score tests and interpretation that clearly tells you what changes to make in your life to be healthier.
I don't want to spend months climbing through this noise and reading every article; I want: "You definitely shouldn't smoke to avoid asthma and eat after 6 to prevent diabetes. If you're interested in how we reached these conclusions, here you go! Everything is absolutely clear and transparent!"
If you know of such providers, please let me know!
Ancestry
With finding my ancestors, I discovered a huge problem - all services that provide the ability to search for ancestors accept not the whole genome, but only part of it obtained through Genotyping.
Therefore, if you have a full genome, you'll need to convert it to the right format. In the appendix, I described how you can convert your WGS genome into genotyping formats for 23andMe, Ancestry, and others.
Genealogical tree for comparing results
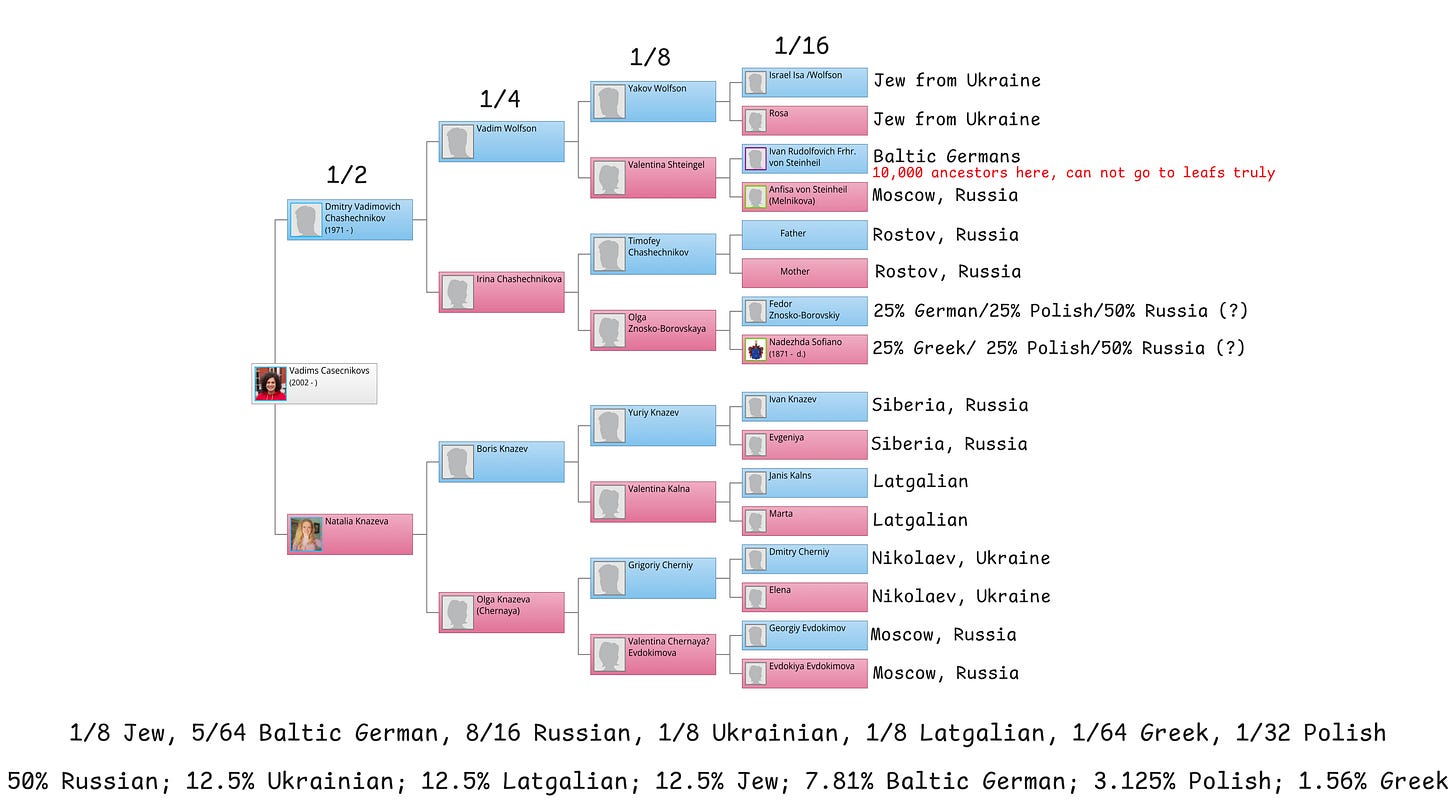
To create some baseline and check services for quality, you can create a genealogical tree.
With the help of my relatives, I created my genealogical tree as deep as I could - about 60 people.
To expand it further, I connected my tree to Geni.com.
With this service, you can connect your tree with the trees of your distant relatives, combining your knowledge about your ancestors.
I was able to find a whooping 9,890 direct relatives (0_0). Mainly due to one ancestor of a cool Baltic German. Deutsche liebe ordnung.
This is what I assume:
50% Russian
12.5% Ukrainian
12.5% Latgalian
12.5% Jewish
7.81% Baltic German
3.125% Polish
1.56% Greek
For most ancestors, the depth doesn't exceed 5-6 generations, I assume that if it's not known that a person was nobility or a merchant, then they didn't move much, and accordingly, nationality can be assigned by the city in which they lived.
Haplogroup analysis
Price: free
To get haplogroups, you need to use WGSExtractor, see the appendix.
Since Y-DNA is passed down only from father to son and doesn't mix with maternal DNA, the only way it can change is through single mutations.
We can trace people whose Y-DNA is similar to ours and assume that we shared a common father of father of father ... of father.
The same story applies to mitochondrial DNA, but we're looking for the mother of mother of mother ... of mother.
Y DNA
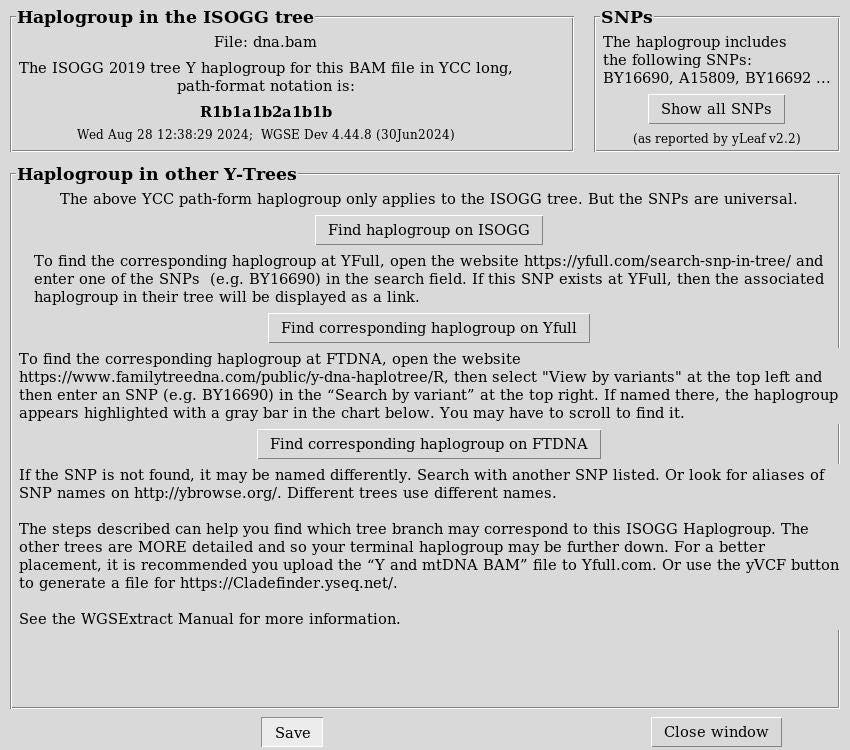
My Y haplogroup is R1b1a1b2a1b1.
https://isogg.org/tree/ is a database of all Y haplogroups.
We find our common haplogroup in it:
The image shows how haplogroups work.
We find the name of our haplogroup - A15807.
We search for it on this site:
https://discover.familytreedna.com/y-dna/R-A11711/story
People who share the closest known common great-grandfather with me are from Belarus, Ukraine, and Poland:
Comparison with baseline
My grandfather was indeed a Jew from Ukraine.
Considering that people with this haplogroup in Eastern Europe make up less than 1%, this could indeed be a Jewish haplogroup.
MtDNA
My MtDNA is H49a.
We search on the same site, but now for MtDNA:
https://www.familytreedna.com/public/mt-dna-haplotree/H;name=H49a
People with whom I share the closest common mother of mothers are currently in Scandinavia and America.
Comparison with baseline
My mother's mother and so on were from the Moscow region of Russia. They didn't have a predisposition for mobility.
It doesn't align with my genealogical tree. But my tree only goes back 5 generations.
The distance to Finland is about one and a half thousand kilometers.
GedMatch
Link
Price: Free
Here I found an amazing guide on how to use GedMatch to get the best results.
Please read it.
I won't be able to explain it better.
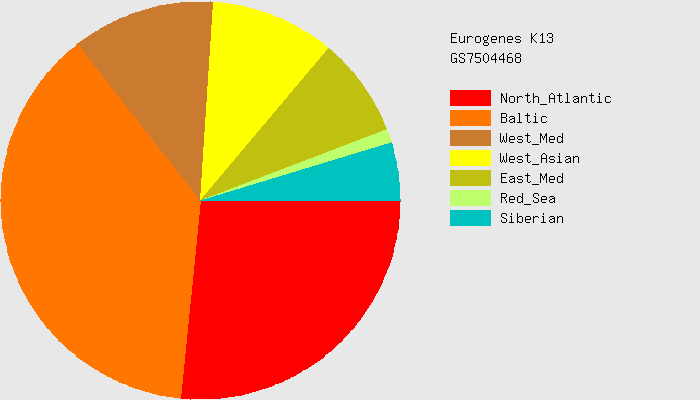
Here's an example for me, which I got from the Eurogenes K13 test.
Baltic 37.7%
North Atlantic 26%
West Med 11%
West Asian 10%
East Med 8%
Sib 2%
Comparison with baseline
For Eurogenes K13, I found a map for interpretation - https://genealogical-musings.blogspot.com/2018/04/eurogenes-k13-maps.html
One could stretch the truth and say that this potentially aligns with my genealogical tree. But according to my analysis, I should be 37.5% Russian from rural Russia, which is hard to count as North Atlantic or even Baltic guys.
This test raises some doubts, but we can assume that the test has some meaningful basis.
Admix
Link
Admix has the same functionality as GedMatch above, but it's a CLI application.
For those who prefer not to share their DNA with various services left and right.
MyHeritage
Link
Price: 35€
You upload your template to MyHeritage
Comparison with baseline
Unlike GedMatch, MyHeritage doesn't stand up to scrutiny.
Where did 8.6% Irish come from???
There are Scots and Irish in the genealogical tree, but they're there for one millionth of all DNA at the level of error. There shouldn't be any 8.6%.
General conclusion on ancestry
Having an approximate understanding of your genealogical tree, we can confidently say that genealogical tests have a significant component of horoscopes for boys.
I assume that haplogroups have high accuracy, but autosomal DNA is tricky, especially for analyzing probabilities below 10%.
If there's a quality WGS Ancestry test out there - please let me know.
ML speculation
My assumption is that these guys are trying to geographically cluster mutations in a space with a small number of dimensions.
If you end up exactly in one cluster or between a couple, you can ± accurately say who you belong to, but if you start to be in a strange position, ML starts to fail.
I assume this is only possible if the dimensions by which clusters can be separated are extremely few.
And if they're extremely few, then we're not that different.
After all, as is known, two orangutans of the same species in the same forest will differ genetically more than the two most different humans on the entire Earth.
I would really like to take a WGS Ancestry test and look at the results, let me know if someone is already doing this.
Capitalist speculation
An alternative hypothesis is that the tests themselves have capitalist incentives.
Users want to get non-obvious exotic relatives.
Moreover, users want the most accurate answers, which forces software creators to say, not "Somewhere in Western Europe," but "Ireland."
Traits
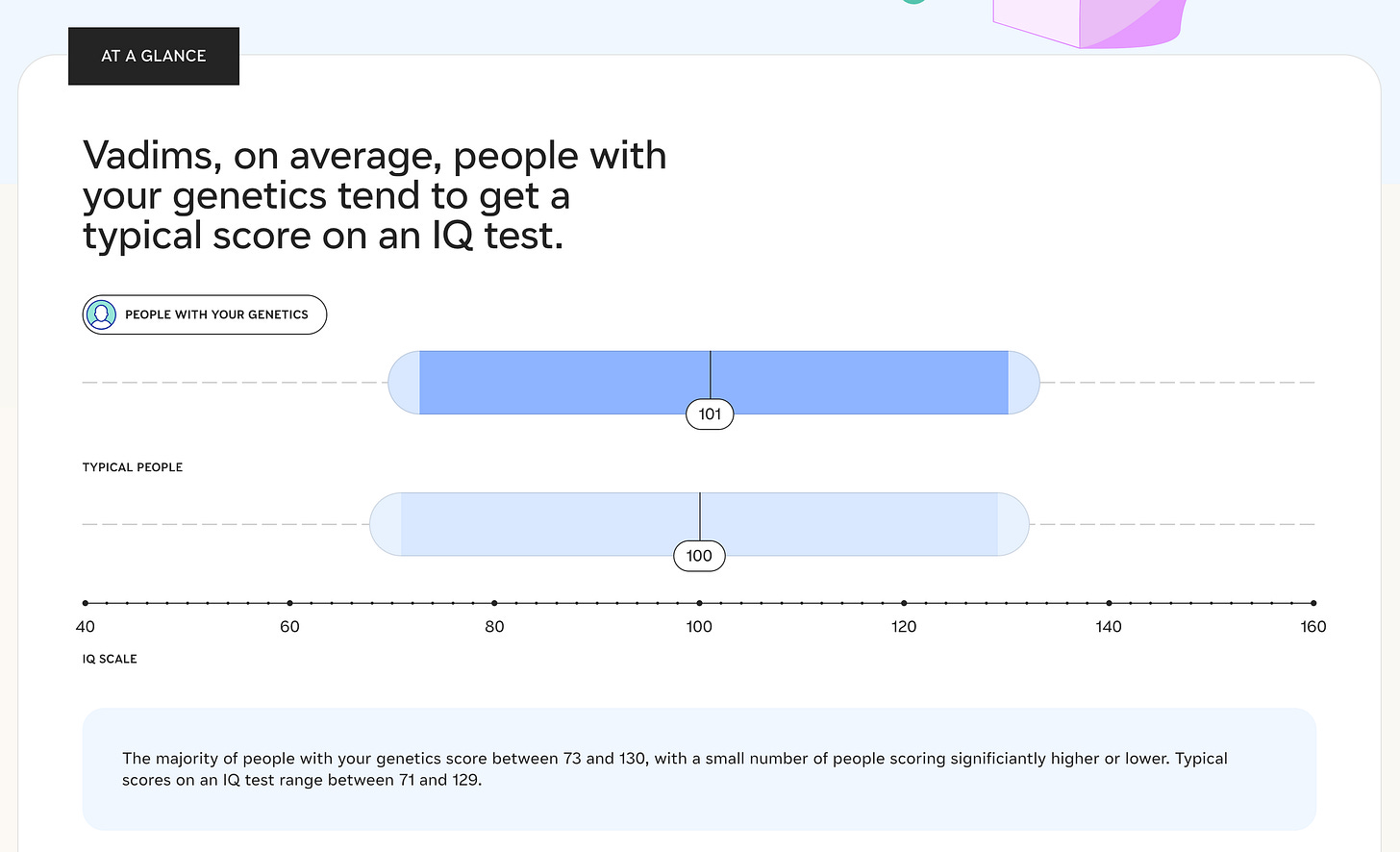
Nucleus IQ
An IQ test only tells you how well you can solve an IQ test and tasks similar to it. It says very little about intelligence.
My IQ according to the Nucleus test is 101 IQ, which, considering the Flynn Effect, is below average.
Two weeks ago, each member of my family took an IQ test. Each family member's test was outside the range indicated by Nucleus (71-129).
There could be two reasons:
The Nucleus genetic test is not accurate
The genetic component plays only a small role in the IQ test, with the cultural component playing a huge role
It's basically a horoscope for boys.
GenomeLink
Link
Price: $14/mo
GenomeLink is one of many providers that try to predict your traits based on your genome.
They offer a huge number of traits, currently around 300 and constantly adding new ones.
Here's an example of what the traits look like:
If you click on them, a link to the source appears:
More than half of the sources are questionable:
Scientific reliability 1/4, population Mexican.
GenomeLink conclusion
GenomeLink clearly shows a strong business incentive to constantly add new traits to the platform, as the payment system is subscription-based.
Because of this, low-quality articles are added for populations that don't correspond to my population.
Moreover, they don't report quantitative results anywhere, how much I differ from the average person in the population. I suspect this might be because the differences are actually fractions of a percent and much smaller than cultural deviations.
The site didn't convince me that these traits significantly affect my life.
I couldn't draw any conclusions after studying this site, I saw white noise.
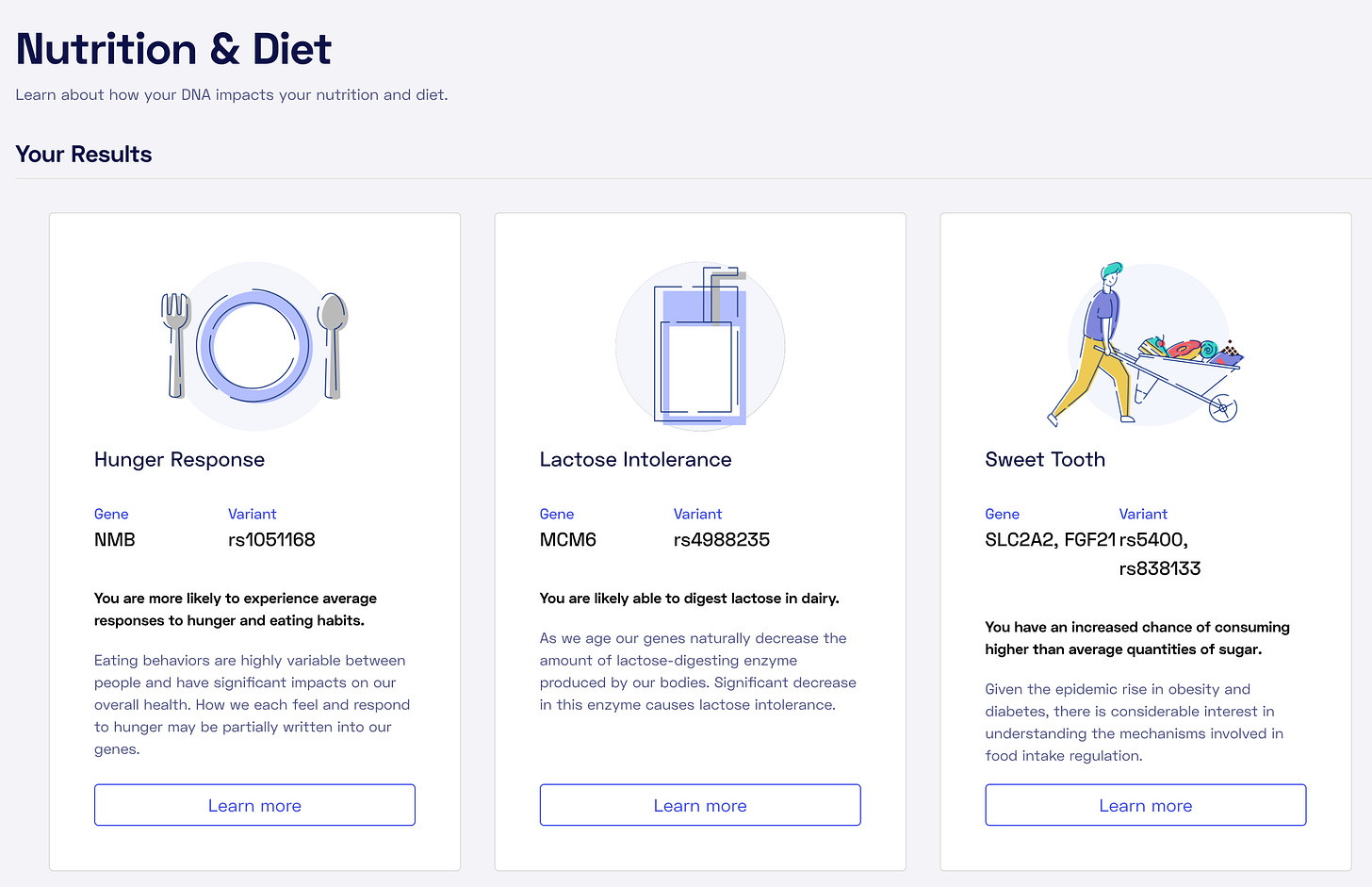
Nebula
Nebula also provides traits in four categories:
Nutrition&Diet
Appearances&Hormones
Behavior&Perception
Body&Athleticism
Here's an example:
For each trait, there is an explanation of the variations used and a link to the article:
Unfortunately, there is no simple quantitative explanation of how much this genetic mutation affects my behavior, so despite the other niceties of the Nebula service.
Traits are a horoscope for boys.
Conclusion
Health
After taking the DNA test, I wanted to receive clear signals - what should I change in my life to be healthier, feel better, and be happier.
Providers worth spending time on:
Nucleus
GeneVue
Nebula
GeneticHealth
I would like to have a service that immediately provides a call-to-action.
Too many services trading in white noise.
Ancestry
Ancestry left mixed feelings, I don't think I learned much new about my ancestors.
I hope to find WGS tests that would give me more accurate data.
Traits
Traits are a horoscope for boys. 0% signal, 100% noise. I couldn't make any conclusions, unfortunately, I wasted money.
I want more signal, more WGS-based analyses, to understand how much a specific variation affects my life in numbers!
Appendix
How to generate 23andMe, Ancestry files from WGS
If you don't have a BAM file but have FASTQ, you'll need to know how to use CLI
If you have a BAM file, congratulations, it will be easier for you!I recommend doing everything on FASTQ
Found an awesome picture on the internet:
WGS Files 101
The genome is read from sample DNA using a Sequencer and we get fastq files.
We can align these fastq files to a template human genome and get a bam file.
To make the data take up less space, we can store only those mutations that differentiate our genome from the template genome. This file is called VCF.
We usually look at variations based on the VCF file - what differs in a person from the normal genome and draw conclusions about whether the mutation is pathogenic or benign.
VCF is usually used for medicine.
Genotyping 101
In genotyping, we read not the entire genome, but only a certain number of genes that interest us. The output of these genes is usually stored in a regular TXT file, as there aren't that many of them.
Genotyping is often enough to make genealogical conclusions.
How to generate genotyping files
Nucleus returns two formats:
1 VCF
16 FASTQ files
Fastq files will have names like:081824-WGS-C3060871_S34_L003_R1_001.fastq.gz
L003 - this is which lane (sample) of our DNA is currently being sequencedR1 - this is in which direction sequencing is currently happening, forward or backward.
Download necessary programs
For converting FASTQ to BAM:
minimap2 (apt get, brew)
samtools (apt get, brew) tutorial
Needed by everyone:
WGSExtract link
Prepare FASTQ files
Concatenate all lanes with R1 together.
Concatenate all lanes with R2 together.
The L's should be arranged in the same sequence in R1 and R2.
The commands will look something like this:
cat 081824-WGS-C3060871_S34_L001_R1_001.fastq.gz 081824-WGS-C3060871_S34_L002_R1_001.fastq.gz 081824-WGS-C3060871_S34_L003_R1_001.fastq.gz 081824-WGS-C3060871_S34_L004_R1_001.fastq.gz 081824-WGS-C3060871_S34_L005_R1_001.fastq.gz 081824-WGS-C3060871_S34_L006_R1_001.fastq.gz 081824-WGS-C3060871_S34_L007_R1_001.fastq.gz 081824-WGS-C3060871_S34_L008_R1_001.fastq.gz > R1.fastq.gz
cat 081824-WGS-C3060871_S34_L001_R2_001.fastq.gz 081824-WGS-C3060871_S34_L002_R2_001.fastq.gz 081824-WGS-C3060871_S34_L003_R2_001.fastq.gz 081824-WGS-C3060871_S34_L004_R2_001.fastq.gz 081824-WGS-C3060871_S34_L005_R2_001.fastq.gz 081824-WGS-C3060871_S34_L006_R2_001.fastq.gz 081824-WGS-C3060871_S34_L007_R2_001.fastq.gz 081824-WGS-C3060871_S34_L008_R2_001.fastq.gz > R2.fastq.gz
Make sure to specify your own FASTQ files!
Installing WGSExtract and downloading the template genome
After downloading the WGSExtract archive from the website, run the install script for your OS.
Launch Library.command and download hs37d5 (NIH). In my case, it was number 5.
This genome will be located in the folder: /WGSExtractv4/reference/genomes/hs37d5.fa.gz
FASTQ to BAM
minimap2 -t 16 -a -x sr /src/WGSExtractv4/reference/genomes/hs37d5.fa.gz /src/R1.fastq.gz /src/R2.fastq.gz | samtools fixmate -m - - | samtools sort -@16 -T /tmp - | samtools markdup - - | samtools view -@16 - -o /src/finals/hs35d5/dna.bam
Converting FASTQ files to BAM.
In the script above, you need to specify:
Your concatenated *.fastq.gz files.
The path to the reference genome with the *.fa.gz extension
The location where the script will save the *.bam file
Processing will take up to 9 hours. Time to brew a cuppa!
BAM to 23andMe
Launch WGSExtract.*, a window with a menu will open
Select the BAM file,
Once the BAM file is loaded, click on
Indexand thenSortGo to the Extract Data tab, click on MicroarrayRaw and choose which genotyping formats you want to create. I recommend creating all of them, as the services that accept them can be a bit unpredictable
(!Bonus) Discovering Mitochondrial and Y haplogroups
In the Analyze tab, select Determine haplogroups:
Y Chromosome
Mitochondrial DNA
Here is my Mitochondrial DNA haplogroup:
And Y Chromosome haplogroup:

















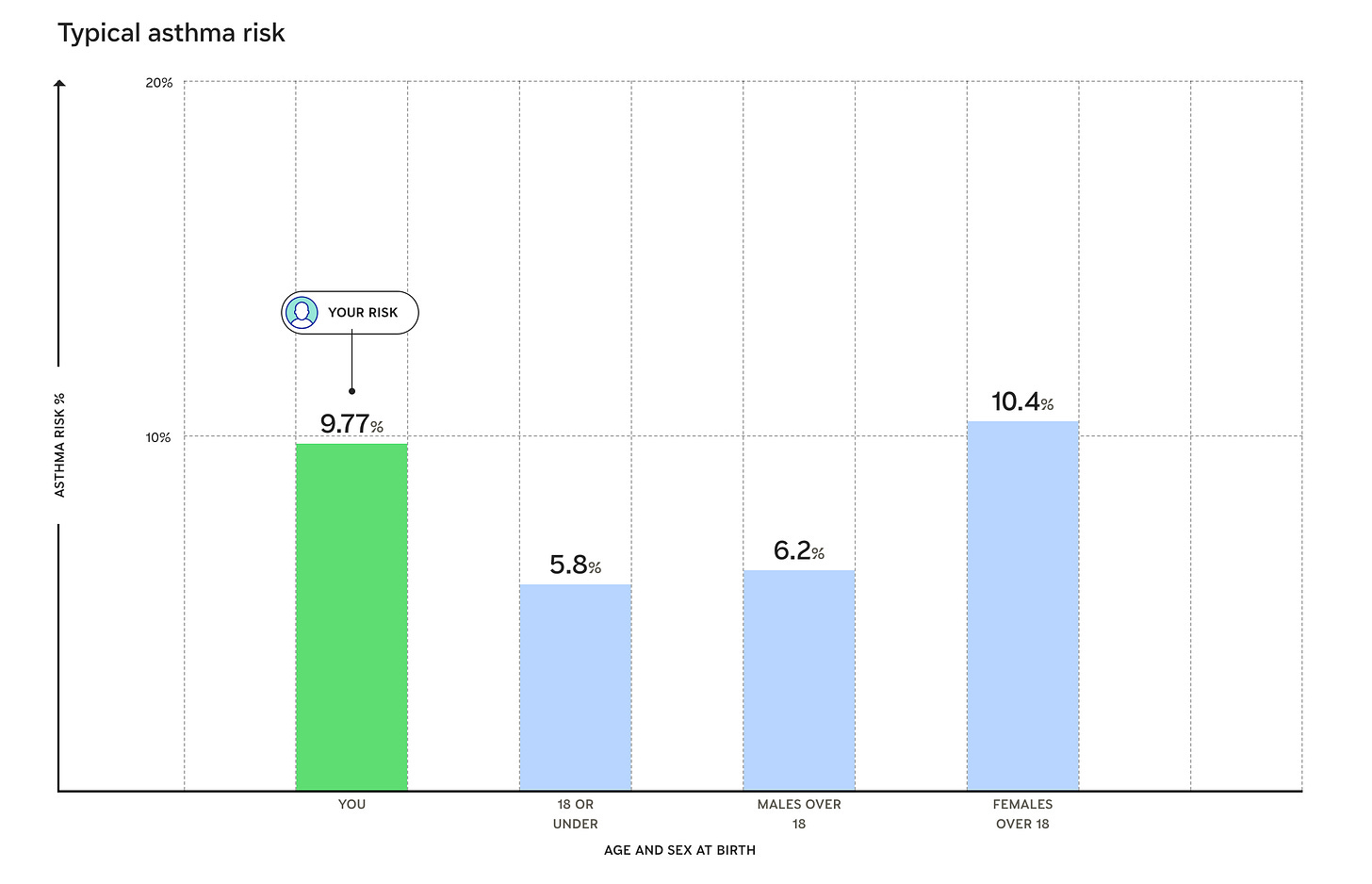























Great post! I’m planning to get my WGS this Cyber Monday. I have two questions:
Is it better to pay for analysis/reports (like Nebula or Sequencingdotcom) or just get the raw BAM file and analyze it myself? Sequencingdotcom’s $299 sale doesn’t include cardiovascular insights, and the $399 option is close to Nebula’s and Nucleus’ prices. Some say Nebula has deeper WGS reports than Nucleus—any thoughts?
Nucleus uses the Illumina NovaSeq X, while Nebula uses MGI T7/T10. Someone in the Personal WGS group mentioned Nebula’s T10 had ~52x read depth (WGSE Beta v4.44.5). What was your read depth from Nucleus?
Each service for ancestry is good at certain things with certain data sets. Like MyHeritage is the best for EU dna matching and jewish ethnicity, LivingDNA is the best for british isles specifics, http://Ancestry.com is best for US dna matching, etc. You might have some irish/scotish in your family tree because something sneaky happened for example, or there are some edge cases you could double check it with LivingDNA. I did for myself since I have a lot of british isles DNA. Ethnicity estimates are also just that, estimates. Very fuzzy in practice.
I also have a bunch of french ancestry, but it didn’t show up, and we realized it probably came from Brittany, which is a genetic exception to the rest of France for example, if you look closely at the blobs they tend to also include Brittany. I think it’s called Brittany due to some invasion or they were originally from Britain.
I also got a bunch of other family members sequenced and it lets you determine which came from what line because they tend to concentrate. Like I got my south asian genetics from my maternal grandmother after sequencing my mother and grandmother with myheritage.
Also in the article, I think you call genetic lifehacks genetic health or genetichealth a few times.
https://www.youtube.com/watch?v=bH9UZoXH-Rw